Over 10 million deaths per year. That's the global death toll
experts predict from drug-resistant infections by 2050 without new ways to
combat these microbes. Unlike most disease areas, where new and improved
drugs are being discovered and developed every year, bacterial infections are
still largely treated with classes of antibiotics that were discovered over 50
years ago. Many of these drugs were found by screening microbes that live in
the soil, which has proven to be a successful strategy for obtaining a vast variety
of chemical compounds. However, most microbes in soil cannot be grown in the
lab, leaving large gaps in our ability to study them and identify potential new
antibiotics. Recent advances, however, have helped overcome this problem and lead
to the identification of the first new classes of antibiotics in 30 years.
 |
The iChip culture method.
Al Granberg. https://www.the-scientist.com/
?articles.view/articleNo/44049/title/Cultural-Riches/ |
In 2015, a team of scientists at Northeastern University isolated a new antibiotic using an ingenious method for growing soil microbes with the iChip, a device with wells for bacteria that are separated from a natural environment (like soil) by a diffusion membrane with tiny pores that allow the transfer of nutrients without allowing the bacteria to leave the well. Using this method, researchers were able to isolate new chemical compounds from these bacteria and identify teixobactin. Upon further testing, they found that teixobactin was highly effective at killing several types of bacteria in culture, including Mycobacterium tuberculosis and methicillin-resistant Staphylococcus aureus (MRSA), by interfering with the ability of the bacteria to build cell walls. Teixobactin was also shown to be effective at clearing infections in mice. Although teixobactin itself is difficult to produce, a group at the University of Lincoln, UK, recently synthesized a much easier-to-make version that maintains potency and could be used for commercial production.
In 2017, another new class of antibiotics was discovered at
Rockefeller University using just bioinformatic analysis of DNA extracted from
the environment; this group didn’t even have to grow the bacteria in the lab.
They based their strategy on the knowledge that there is a family of
calcium-dependent antibiotics. By isolating DNA directly from environmental
samples and searching for sequences that featured the known calcium-binding
signature, they could identify genes that potentially encoded new calcium-dependent antibiotics. They then transferred these DNA sequences into
bacteria that can be cultured for further study. The new antibiotic class they
found, the malacidins, also interferes with the ability of bacteria to properly
form cell walls. Malacidins were shown to clear MRSA infections in the cut
wounds of rats. Importantly, even after 3 weeks of exposure to the drug, there
was no sign of resistant bacteria.
This finding suggests that the mechanism of action for the malacidins is one
that cannot be circumvented easily by the bacteria, which bodes well for their
use against multidrug-resistant pathogens.
A third novel class of antibiotics was just described earlier this
month in the journal Molecular Cell by
researchers from the University of Illinois at Chicago and the biotech company
Nosopharm. This class, called the odilorhabdins, was isolated from a bacterium
that lives in a symbiotic relationship with a nematode worm in soil. This bacterium secretes a number of compounds that help the nematode colonize and
kill insects and keep the insect carcass from being invaded by other bacteria
or fungi. The odilorhabdins exert their antimicrobial activity by interfering
with bacterial protein production. While several other antibiotics also target this
process, the odilorhabdins bind to a unique site on the bacterial ribosome (the part of the cell responsible for making proteins);
this means that bacteria that are resistant to other antibiotics that interfere
with protein production will not be resistant to the odilorhabdins. When
tested for their ability to kill several pathogenic bacteria in culture, the
odilorhabdins were highly effective. One of the odilorhabdins, NOSO-95179, was
also tested in mice and could significantly reduce Klebsiella
pneumoniae septicemia and lung infection.
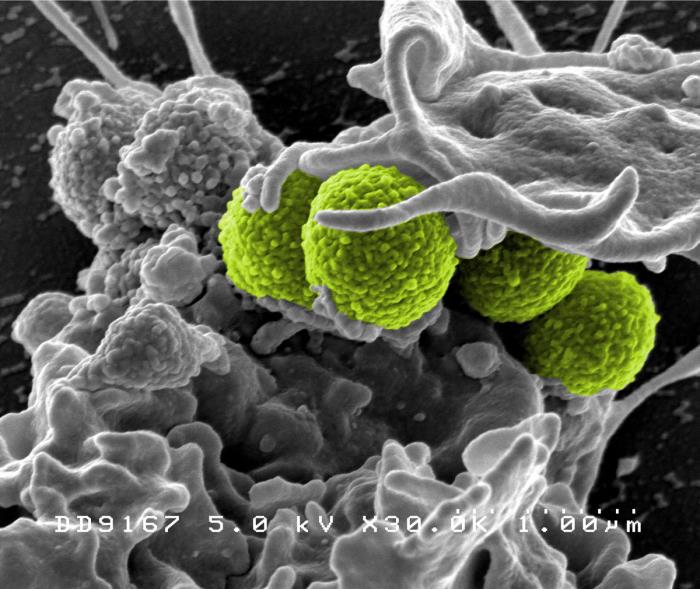 |
MRSA (green) being enveloped
by a white blood cell.
CDC's Public Health Image Library.
Image # 18126; photo credit: NIAID. |
While these new antibiotics are still in the early stages of
development and are at least 6-10 years from being available for use in people,
the rapid identification of multiple new antibiotic classes after a decades-long
“discovery void” provides hope. The new
techniques for culturing soil bacteria, along with bioinformatic approaches
that avoid this step all together, offer new avenues for drug discovery. Additionally, there is ongoing work to re-investigate previously sidelined drug candidates and modify existing antibiotics to improve their efficacy and overcome
resistance. No matter the approach, the road to develop a new antibiotic is long and expensive, making it impossible for academic research labs to go it alone. Luckily, pharmaceutical companies are beginning to get more involved. In 2016, the Antimicrobial Resistance Industry Alliance presented the “Davos
Declaration” at the World Economic Forum. Nearly 100 companies signed
the declaration, pledging to support the research and development of new antimicrobials and improve access to current and future treatments. With this renewed commitment from the pharmaceutical industry, which had largely been turning away from antimicrobial development in recent years, the financing and man-power to produce these new antibiotics might just be available. While nothing short of a strong, concerted effort to deal with drug resistance will allow us to avoid the looming projection of 10 million deaths per year, the current pace of advancement bodes well for our ability to rise to the challenge.

